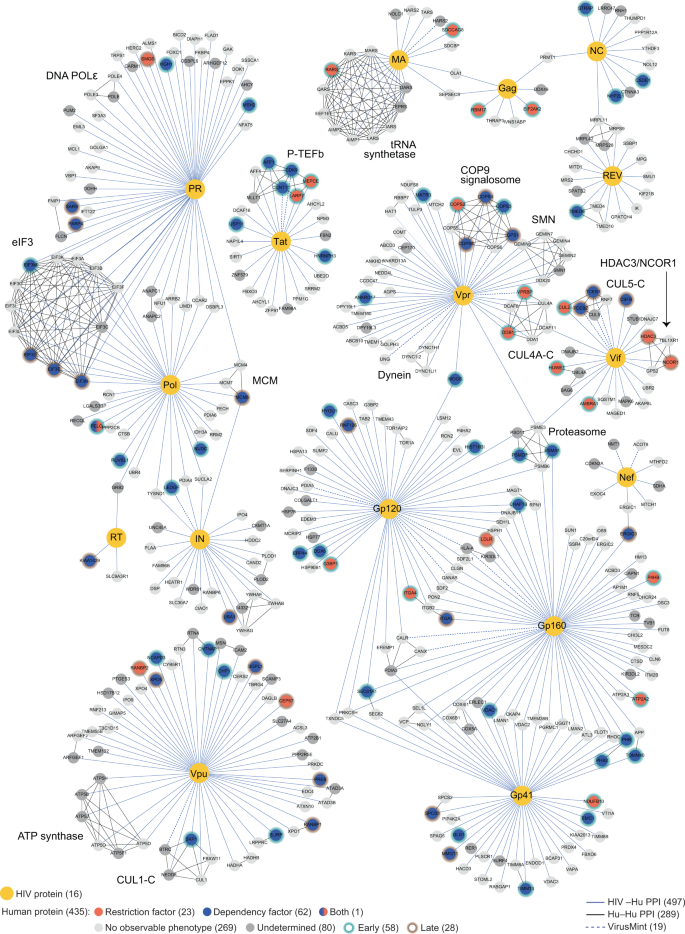
Like all viruses, human immunodeficiency virus (HIV) is unable to replicate on its own. To replicate, it hijacks the molecular machinery that’s found within the host cells it infects. In a study published April 1, 2022, in Nature Communications, a team from the Quantitative Biosciences Institute (QBI) at the University of California San Francisco (UCSF) and the Gladstone Institutes reported that they had created a functional catalogue of these hijackings, providing a new window into HIV biology.
To do this, the team used CRISPR-Cas9 gene editing to create a functional map of the interactions between HIV and human T cells isolated from blood. Of the 86 proteins the researchers identified that alter HIV infection, about half were previously unknown. The investigators say this study opens the door for new research into these interactions and for ultimately developing new drugs to block them. The research also provides important validation of using CRISPR technology to study virus-host interactions directly in human T cells, a new area of research.
“Existing antiviral drugs have been incredible at decreasing the replication of HIV in the body, but these drugs have a number of shortcomings, including the fact that people are tied to taking them for the rest of their lives,” says co-first author Judd Hultquist. “For us to find a functional cure, we need to better understand how the virus is replicating in the human body.” Hultquist, who is now an assistant professor in the Feinberg School of Medicine at Northwestern University, conducted the research while he was a postdoctoral fellow in Nevan Krogan’s lab at UCSF. Krogan, one of the paper’s senior authors, is Director of QBI.
For this research, the investigators focused on protein-protein interactions. Specifically, they looked at the 426 human genes that encode proteins that had previously been demonstrated to interact directly with HIV. They used CRISPR-Cas9 to systematically knock out each gene and then looked at the effect each gene deletion had on virus replication. They found that the absence of some genes made replication go up, while others made it go down.
“This research allowed us to map very specifically how HIV interacts with all of these human proteins,” says co-first author Joseph Hiatt, an MD/PhD student in the labs of Dr. Alexander Marson and Dr. Nevan Krogan. “When you find these new host factors, it points to avenues for new classes of drugs. It’s an incredibly powerful and valuable approach to learning how HIV hijacks cells.”
The investigators focused on this research project for more than six months, often working day and night. Using T cells obtained from human donors’ blood, they painstakingly knocked out each gene and studied how it affected HIV. “This work required so much vision from Nevan and Alex to gather all these resources and bring together these separate lines of technology in a way that can really shed light on an important human condition,” Hiatt says. “This work provides an approach for studying protein-protein interactions more broadly, including in cancer and other types of infections.”
“For the past two years, the world has been fixated on the ongoing COVID-19 pandemic, while other epidemics have faded into the background,” Hultquist says. “But there are currently 38 million people living with HIV, including 1.5 million new infections that occurred in the past year. We have the technology to better answer basic questions about HIV biology and we need to use them to develop new therapeutic strategies. We do not want HIV to become the forgotten epidemic.”