Trentini et al.
Arginine Phosphorylation Marks Proteins for Degradation by a Clp Protease
Nature 2016, 539, 48-53.
http://www.nature.com/nature/journal/v539/n7627/full/nature20122.html
Reviewed by Fatima Ugur, CCB, Fujimori Lab.
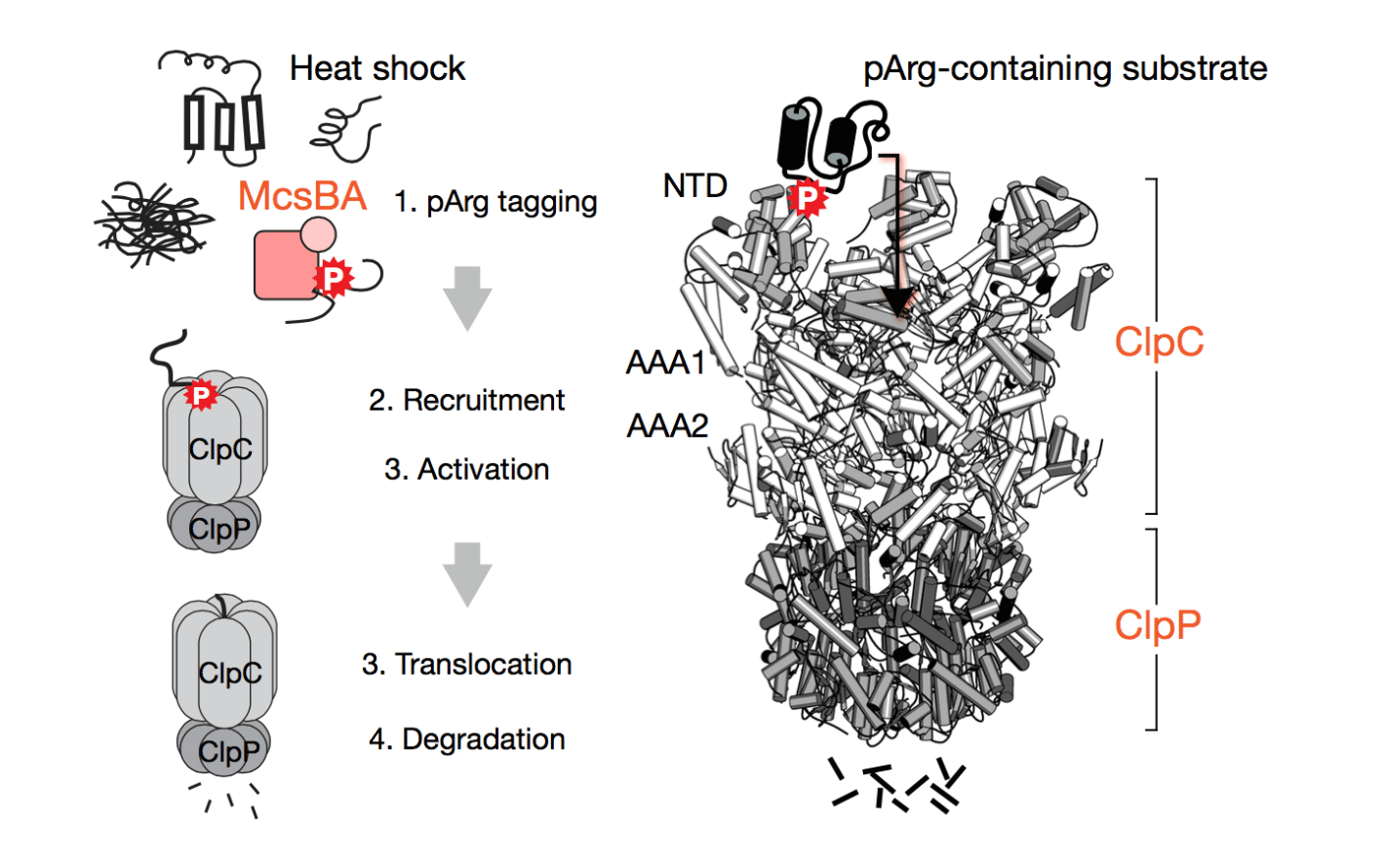
In this paper, they identify arginine phosphorylation as a new post-translational modification (PTM) to target proteins for degradation by the ClpCP proteolytic system in gram-positive bacteria. Arginine phosphorylation is a unique PTM utilized by gram-positive bacteria in the stress response pathway and is catalyzed by the arginine kinase McsB. McsB is known to phosphorylate arginines on approximately 100 substrate proteins, one of which is the repressor CtsR of heat shock genes (including clpP and clpC, the proteolysis system of gram-positive bacteria), linking arginine phosphorylation with protein degradation during the heat shock response. The ClpCP system degrades protein substrates through the ATPase ClpC which unfolds and translocates substrate into the protease ClpP. An adaptor protein MecA induces degradation of substrate proteins by recruiting the substrate to ClpC. In this paper, they investigate how McsB serves as an adaptor protein of the ClpCP system, how phosphoArg affects substrate protein degradation, and how phosphoArg is important in responding to heat stress.
They identify phosphoArg marks on ClpP substrates and show that phosphoArg catalyzed by McsB induces substrate protein degradation by ClpCP without the presence of other adaptor proteins in vitro. Binding of phosphoArg by ClpC is through a unique phosphorylation recognition site and binding induces ClpC ATPase activity. phosphoArg-mediated protein degradation by ClpCP was shown to be important for B. subtilis to survive heat stress. They propose a model of phosphoArg-dependent protein degradation by ClpCP for the degradation of unfolded proteins after heat stress. They suggest that arginine phosphorylation might tag unfolded proteins during the heat shock response and might prevent proteins from aggregating, while targeting them for degradation. However, they do not perform the biophysical experiments to assess these effects of phosphoArg on substrates, in addition to assessing the degradation of endogenous phosphoArg substrates by ClpCP. Overall, it is interesting that a bacterial 'equivalent' of ubiquitin, a reversible PTM, has been identified to target proteins for degradation. However, more work is needed to assess the biological role of arginine phosphorylation and whether phosphoArg mediated degradation is a more global pathway of protein degradation in bacteria.