The human genome has captured popular imagination since the first draft sequence nearly 20 years ago and this has led to a better understanding of disease, widely benefitting fields ranging from molecular medicine to human evolution. Yet for decades, a key driver of the genome’s ability to specify cell identity was misunderstood.
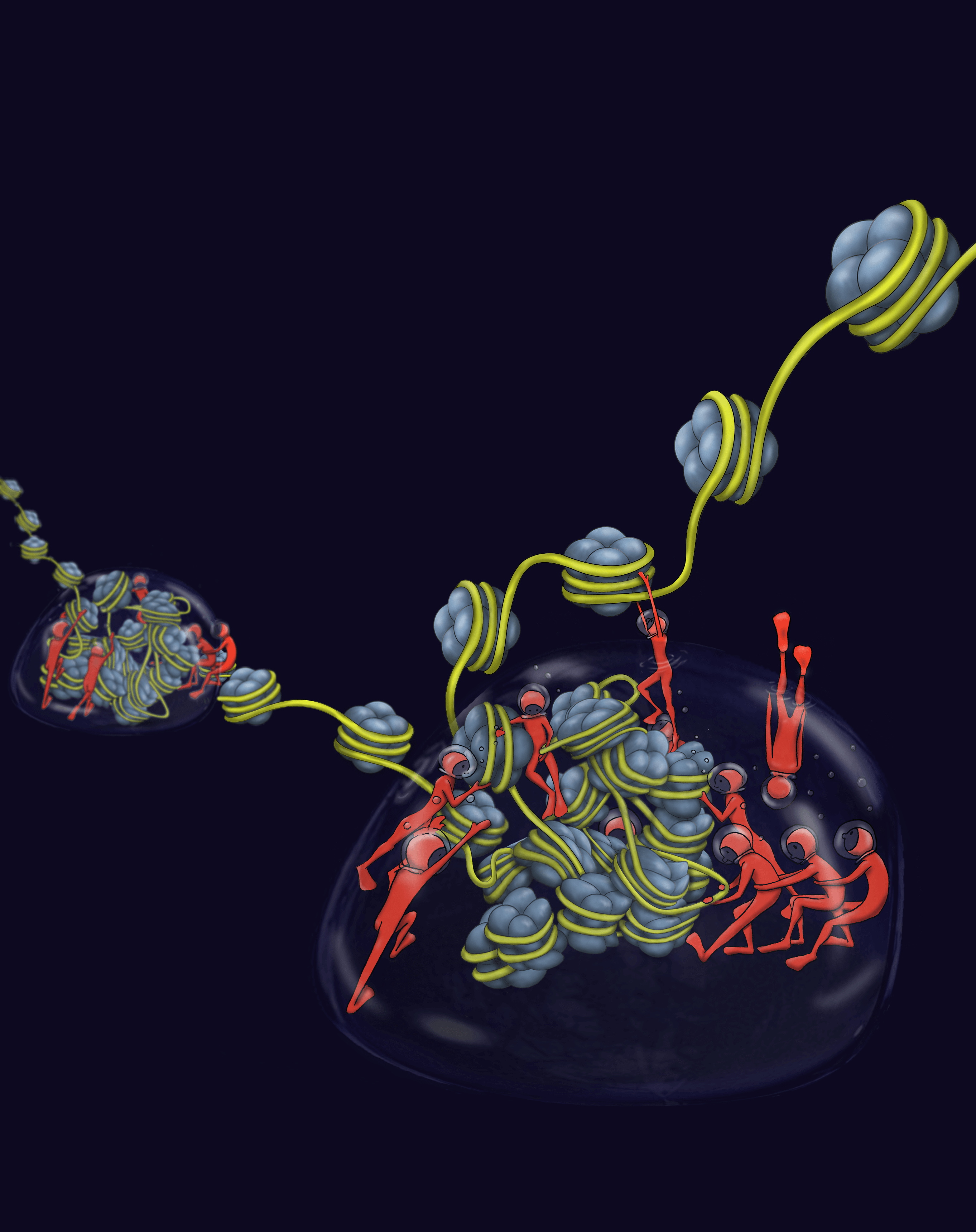
To exert its coding power, the human genome must be appropriately packaged into structures called chromatin that dictate which genes can be “read” and which cannot. This packaging helps determine whether a stem cell becomes a heart cell or a brain cell and whether a cell remains normal or becomes cancerous. In textbooks, the fundamental building block of chromatin, a nucleosome, is compared to a wooden spool that organizes thread. This is because a nucleosome is made of eight histone proteins that are thought to snap together like lego pieces to form a rigid structure around which strands of DNA are wrapped. Multiple nucleosomes can then be compacted together by the action of the protein HP1 to form structures termed heterochromatin. Heterochromatin is particularly effective at preventing the underlying genes from being read. In textbooks heterochromatin is often described as a static and “locked down state”.
A 2017 study from the labs of senior authors, Geeta Narlikar, PhD, professor of biochemistry and biophysics at UCSF, and John Gross, PhD, professor of pharmaceutical chemistry at UCSF, challenged the textbook model for nucleosomes. This study demonstrated that nucleosomes are not rigid but instead can locally change their shape to make genes accessible for reading. Another study from the Narlikar lab in 2017 showed that HP1 proteins compacted DNA in phase-separated liquid condensates, very much like oil and water. This finding challenged the notion that heterochromatin is a static and “locked down” state.
Ever since, an array of new questions has risen to the surface, further making way to key discoveries that broaden our understanding of epigenetics. In particular, researchers have been asking how much and how often nucleosomes change their shape and how such shape-shifting influences genome packaging.
A new study, published in the journal Nature on October 16, 2019, further exemplifies the idea that treating and studying diseases caused by defects in genome packaging requires re-imagining what nucleosomes and heterochromatin look like.
Thinking of nucleosomes as deformable and heterochromatin as a dynamic liquid-like compartment led Serena Sanulli, PhD, a postdoctoral researcher in the labs of senior authors Narlikar and Gross, to ask what exactly happens to nucleosomes in heterochromatin. Dr. Sanulli found that the HP1 protein increases the accessibility of histone residues buried deep into the nucleosome by opening up its core, essentially reshaping the nucleosome’s interior. Sanulli called this “dancing nucleosomes.”
“You ask yourself how to make sense of it…” said Narlikar, “One way is to imagine you have a mug and are trying to pack many beads into it to see how compact it can get. If the beads are rigid wooden beads (like the spools) there are only a certain number of them you can pack. If the beads are squishable, you can fit many more beads in there.”
The eight histone proteins inside the nucleosome essentially can now be perceived more like malleable playdough balls, rather than rigid Lego blocks that snap together. In order to fit in a compacted space, histone proteins change shape and adapt, which requires flexibility, adjustment, adaptation, and movement, very much like in a dance.
This suggests that altering the shape of the fundamental building block of genome packaging can have large effects on processes ranging from gene regulation to genome organization.
These findings also offer a new view of how to interpret histone modifications that are found buried deep inside the nucleosome. These modifications could directly control shape shifting by nucleosomes and thereby enable or inhibit the extreme compaction observed within chromosomes.
Sanulli said she began the project with very basic knowledge, having come from a different field where she studied stem cells in mice development. “I approached the project with a different lens and focused on it at a very atomic level. I realized how challenging it was and how very few labs around the world were doing it,” she said.
Although it took time to get results, Sanulli said that trying complementary approaches eventually led to the big reveal. “All at once it started working. We saw some very crazy conformational changes, implying new shapes of the nucleosomes. We were surprised and did not expect it. But because we had three independent methods that were pointing at the same thing, we had to believe it,” she said.
Co-senior author, Gross, said this project is an example of pure curiosity-driven science. “We started this project as a bit of a dare, thinking ‘we can do this, let’s just look.’ Serena had a lot of confidence. Instead of pushing back, she got excited about how it could challenge the way one looks at chromatin. She was very adventurous and bold and, in a way, trusting that there would be something really rich that she could get from this. That is certainly all true,” he said.
Narlikar described this research as being the beginning of future inquiries. Scientists need only be limited by their imagination in developing and testing models for how shape-shifting nucleosomes control genome activity.
Because genome packaging controls cell identity, defects in genome packaging can cause a cell to have an identity crisis. For example, defects in genome packaging could confuse brain cells to behave like cancer cells or cells involved in diseases like Parkinson’s disease. Thus understanding the basics of genome organization has implications to various areas, from cancer to neurodegeneration.
“A colleague of mine at another university found that several cancer related mutations, are within the histone proteins in the spool and could affect the ability of nucleosomes to change shape. Now we are getting ready to test the mutations and ask ‘do those mutations make nucleosomes more or less breathable?’” said Narlikar.
Ultimately, this finding is one of many critical pieces of a puzzle that helps scientists see how the genome is packaged and how this packaging regulates health and disease.
“The cars and planes we see today arose from many inventions building on one another, … we and our field are figuring out how Nature works through multiple discoveries that build on each other,” said Narlikar.
Other authors on the paper were Michael Trnka; Venkat Dharmarajan; Ryan Tibble; Bruce D. Pascal; Al Burlingame; Patrick Griffin.